2 Institute of Animal Husbandry, Heilongjiang Academy of Agricultural Sciences, Harbin, 150040, China
3 Graduate School of Natural Sciences, Nagoya City University, Nagoya, 467850, Japan
 Author
Author  Correspondence author
Correspondence author
Molecular Soil Biology, 2016, Vol. 7, No. 7 doi: 10.5376/msb.2016.07.0007
Received: 05 May, 2016 Accepted: 01 Jun., 2016 Published: 15 Jun., 2016
Dandan Li, Xiaoqing Huang, Ziguang Liu, Shuang Li, Toshihiro Okada, Yasushi Yukawa, and Juan Wu, 2016, Effect of AtR8 lncRNA Partial Deletion on Arabidopsis Seed Germination, Molecular Soil Biology, 7(7): 1-7 (doi: 10.5376/msb.2016.07.0007)
Long non-coding RNAs(lncRNAs) are longer than 200 nucleotides in length, which largely exist in organism and have various biological functions. AtR8 RNA is a lncRNA transcribed by RNA polymerase III in Arabidopsis. Northern blot analysis indicates that AtR8 RNA is abundant during seed germination. AtR8 RNA partial deletion reduces seed germination under normal culture condition. SA treatment induces AtR8 RNA expression. High concentrations SA treatment decreases seed germination of wild type and AtR8 RNA partial deletion mutant, but the inhibition for AtR8 RNA partial deletion mutant is more obviously. All the results show that AtR8 RNA may involve in arabidopsis seed germination process.
1 Introduction
Long noncoding RNAs (lncRNAs) are more than 200 nucleotides in length, which do not encode proteins and play a role at RNA form (Ben et al., 2009). Research shows the lncRNAs play an important role in dosage compensation, genomic imprinting and X chromosome inactivation and other biological processes in mammals (Huang et al., 2015). The research of plant lncRNAs is still in its infancy, known mechanisms are only found in a few of them. In Arabidopsis, AtIPS1 (Induced By Phosphate Starvation 1) inhibated activity on miR-399 by imitating miR399 target gene PHO2, and it could maintain growth of roots under phosphate starvation (Zhang et al., 2013) ; COOLAIR (cold induced antisense intragenic RNA) regulated flowering time by inhibiting FLC gene expression with transcription interference(Swiezewski et al., 2009); COLDAIR (cold assisted intronic noncoding RNA) made FLC gene epigenetic silencing by histone modifications on H3K27me3, influencing flowering time (Heo et al., 2011); HID1 (HIDDEN TREASURE 1) modulated hypocoty growth through regulating PIF3 (PHYTOCHROMEINTERACTING FACTOR 3) expressing in continuous red light (Wang et al., 2014); APOLO transcribed by pol II and V and responding to auxin, its dual transcription impacted the chromatin topology and expression of its neighboring gene PID (a key regulator of polar auxin transport), thus affected root growth (Ariel et al., 2014); ASCO (Alternative Splicing Competitor) acted with AtSR (nuclear RNA-binding protein plaques) and alternatively spliced mRNA target gene, and it regulated the alternative splicing pattern of NSR and ASCO through intercepting alternative splicing regulator to affect development of Arabidopsis lateral root (Bardou et al., 2014). In rice NK 58S (Nongken 58S, an elite japonica rice variety), a C-to-G mutation of LDMAR (long-day-specific male-fertility– associated RNA) sequence caused methylation at promoter region and inhibited LDMAR expression, as a result of PSMS (Photoperiod-sensitive male sterility) under long-day conditions (Ding et al., 2012).
Seed is a specific organ of plant, which plays an important role in continuation of the species. It is usually consisted of three parts, testa, embryo and endosperm. Depending on the absorption rate, seed germination process is divided into three phases: Phase I (imbibition phase), the rapid absorption process of dry seeds under suitable conditions, Phase II, the absorption rate of seed is to slow down, water content is relatively stable and seed coat begins to break down; Phase III, endosperm ruptures, and radicle projects seed coat, which means germination is completed (Krishnan et al., 2004). Seed germination is influenced by many genes and environmental factors. Early analysis and molecular cloning studies found, the ABI3, CHI, DFR and FUS3 and other genes were related to seed germination (Holdsworth et al., 2008; Giraudat et al., 1992; Shirley et al., 1995). Recent studies have found the extension AtEXPA2 gene specifically expressed in Arabidopsis seed germination stage, and involved in GA-mediated process and increased the tolerance of salt and osmotic stress during seed germination (Yan et al., 2014). Three expansion genes were also found to be expressed in germinating tomato (Lycopersicon esculentum Mill.) seeds. LeEXP4 was expressed specifically in the seed radicle cap, it was related to radicle breaking the seed coat. LeEXP8 was accumulated specifically in the radicle cortex during and after germination. LeEXP10 was massive at an early stage of seed development during the period of rapid embryo expansion. Deficiency of endogenous GA inhibited the expression of LeEXP8 and LeEXP10 in imbibed seeds, but was improved by adding exogenous GA. Tissue localization and expression pattern of LeEXP8 and LeEXP10 revealed their special roles in embryonic and seedling growth (Chen et al., 2001). With the rapid development of ncRNA researches, specifically expressed and highly conserved ncRNAs were gradually found in seed germination, a few of them transcripted by pol II were resolved clearly, such as in Arabidopsis miR156 gene reduced the transcription of SPL13 (Squamosa-promoter Binding Protein-like 13) mRNA, delayed development of post-germination seedlings (Ruth et al., 2010A; Ruth et al., 2010B); miR160 negatively regulated ARF10 (Auxin Response Factors 10) gene expression, playing an important role during seed germination and embryo developmental processes (Liu et al., 2002); miR167 regulated the expression of ARF gene family and influenced auxin signaling, effected on seed germination and the embryo development (Wang et al., 2012). At present, lncRNAs have not been reported at seed germination.
Salicylic acid (SA) is a kind of endogenous hormones and has great physiological effects on plant, such as disease resistance, low temperature resistance, drought resistance, salt resistance, ultraviolet radiation (Wang et al., 2012). The previous research about SA has focused on the action of salicylic acid signal molecule about plant resisting disease reaction (Mariana and Javier, 2011). Recent studies have shown that SA functions differently at the stage of plant growth and development, such as Nishimura found that SA deficiency delayed Arabidopsis leaf senescence (Nishimura et al., 2005), Martinez found that SA deletion delayed Arabidopsis flowering time (Martínez et al., 2004). Seed germination quality is an important agronomic trait impacting on growth and productivity. The roles of gibberellin, abscisic acid and ethylene in seed germination have been widely studied. Studies have shown that the uses of different concentrations of SA either suppress seed germination or improve seed vigor, and the result is controversial (Lee et al., 2010). The mechanism of SA remains unclear about seed germination, and it is widespread concerned currently.
In previous studies, according to the promoter structure and transcriptional activity of pol III, we in silico predicted Arabidopsis genome and found AtR8 RNA, which was transcripted by pol III and abundantly expressed in the early elongation zone of the root, and responded to hypoxic stress environment (Wu et al., 2012). This study analyzes AtR8 RNA expression characteristics under normal culture and SA stress during Arabidopsis seed germination, and investigates the effect of AtR8 lncRNA partial deletion on seed germination. It will lay the foundation for the lncRNAs research at seed germination.
2 Materials and Methods
2.1 Plant materials and growth conditions
Arabidopsis thaliana (L) Heynh accession Columbia (col), Wassilevskija (WS) and an AtR8 RNA partial defective mutant derived from WS. After breaking dormancy at 4 °C for 3 d, the seeds are incubated at 22 °C with 16 h light/8 h dark in a growth chamber. Columbia seeds were used for northern blot, Wassilevskija seeds and the mutant were used for seed germination statistical.
2.2 Stress treatments
After surface disinfection with 70% (v/v) alcohol solution and 5% (v/v) sodium hypochlorite solution, wild type Arabidopsis seeds and defective mutant derived from WS were sown on MS-agar medium supplemented with either 5 µM, 10 µM, 15 µM, 20 µM, 25 µM, 0.05 mM, 0.10 mM, 0.15 mM, 0.20 mM, 0.25 mM SA under 16 h light/8 h dark at 22 °C in a growth chamber. Germination was defined as an obvious appearance of the radical through the seed coat. The seed germination percentages were evaluated every day during the germination tests. Fresh weight and dry weight were measured after 7 days cultivation.
2.3 RNA Extraction
0.1 g samples that were ground to powders in liquid nitrogen were homogenized in 1 mL RNA extraction buffer (45.5% (v/v) phenol, 9% (v/v) chloroform, 0.45% (w/v) SDS, 41 mM LiCl, 2 mM EDTA, 5.9 mM b-mercaptoethanol, 82 mM Tris–HCl, pH 8.2), vortexed 1min. The supernatant was added with one volume of phenol: chloroform: isoamylalcohol (25:24:1(v/v/v)) solution, vortexed 1 min. The supernatant was recovered upper layer, and was added an equal volume of chloroform, and incubated at room temperature. The supernatant was recovered upper layer, and was added with 8M LiCl, and stayed at -20°C overnight. The next day the supernatant solution and 1/4 volume of isopropanol was added, and stayed 30 min at -20°C. The supernatant solution and 3/5 volume of isopropanol was added, and stayed 30 min at -20°C. The precipitate was washed with 75% ethanol. Removed the liquid and the RNA was resuspended in DEPC-treated water, at last stored at -80 °C.
2.4 Northern blot analysis
RNA (8 µg) was separated on a 6% polyacrylamide gel and transferred onto a nylon membrane. After UV cross-link for RNA detection, the membrane was hybridized with DIG-labeled RNA overnight at 42 °C. Washing the membrane twice with 2×SSC/0.1% SDS at room temperature and 0.2×SSC/0.1% SDS. After blocking with 1.5% blocking reagent and treatment of anti-DIG AP conjugated antibody, the blots were detected by using LAS 4000.
3 Result and Discussion
3.1 AtR8 RNA expression characteristics during Arabidopsis development
In order to determine the expression characteristics of AtR8 RNA during Arabidopsis development, RNAs are extracted from dry seeds, germinated 36 h seeds and two weeks seedlings, and hybridized with DIG-labeled AtR8 RNA specificity probe. The results show that AtR8 RNA expression is the highest at germinating 36 h (Figure 1).
3.2 AtR8 RNA expression characteristics during Arabidopsis seed germination under SA stress
We investigate AtR8 RNA expression in germinated 36 h seeds with various concentrations of SA (5 μM, 10 μM, 15 μM, 20 μM and 25 μM) treatment. The result shows that with 5 μM SA treatment, AtR8 RNA expression is significantly inhibited, with 15 μM SA treatment, the expression is significantly induced, and the rest do not significantly affect AtR8 RNA expressing. We further investigate that treated at different times (12 h, 24 h, 36 h and 48 h) with 15 μM SA, and it is found that the expression is the highest when treated with 24 h (Figure 2).
.png) Figure 1 Expression analysis of AtR8 RNA during Arabidopsis development. Note: ds: dry seed, gs: germinating 36h seed, sl: seedling (about 7 days) |
.png) Figure 2 Expression analysis of AtR8 RNA during Arabidopsis seed germination under SA stress. Note: A: Expression analysis of AtR8 RNA under various SA concentrations; B: Expression analysis of AtR8 RNA treated with 15 μM SA at different times |
3.3 Seed germination analysis of wild type and AtR8 RNA partial deletion Arabidopsis under different SA concentrations
The AtR8 RNA partial defective mutant is identified at DNA level using PCR amplification with primer, LBP:5'-CGTGTGCCAGGTGCCCACGGAATAGT-3', LP:5'- TACCTGTTCATCG ACGAATTTAGACCAGA-3';RP:5'- GGCTTAAGTCGGCGTTGCGT-3' (Figure 3A),and it is identified at RNA level using northern blot analysis with DIG-labeled AtR8 RNA specificity probe (Figure 3B). The results show that AtR8 RNA mutant is a partial deletion mutant.
The result shows that, under normal culture condition, seed germination rate of AtR8 RNA deletion is lower than wild type. At the concentration of 5 μM, 0.05 mM, 0.10 mM and 0.15 mM SA stress treatment, both of the germination rate barely differ from normal culture (partial data not shown). When the concentration up to 0.20 mM and 0.25 mM, wild type and AtR8 RNA partial deletion are both inhibited at the aspect of seed germination, but the mutant is more evident. Under different concentrations of SA treatment, the result shows that fresh weight and dry weight of wild type are heavier than mutant under normal culture condition. With the increasing concentration, both fresh and dry weights are significantly reduced, but wild type is higher than mutant all the time. And the result is consistent with seed germination statistics (Figure 4).
.png) Figure 3 Identification of AtR8 RNA mutant. Note: A: DNA level identification; B: RNA level identification |
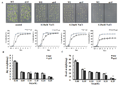 Figure 4 Seed germination analysis of wild type and AtR8 RNA partial deletion under different SA concentrations. Note: A: Under different concentrations of SA treatment, growth conditions and germination statistical analysis of wild type and AtR8 RNA partial deletive mutant after seeds germinating 7 days. B: Fresh weight analysis of wild type and AtR8 RNA partial deletive mutant after seeds germinating 7 days. C: Dry weight analysis of wild type and AtR8 RNA partial deletive mutant after seeds germinating 7 days and dried at 70 ℃ for two days. atR8: AtR8 RNA partial deletive mutant. Data is the result of three repeated experiments. Statistical significance is determined using the Student’s t-test.*represents p < 0.05 and ** represents p < 0.01 |
4 Discussion
Salicylic acid (SA) is a well known endogenous plant signal molecule involved in disease resistance and many growth responses. In normal plant, the content of SA is very low. When pathogen infection occurs, SA increases. Catalase activity is inhibited through SA binding with SABP (salicylic acid receptor protein), and it leads to the accumulation of antioxidants by improving the level of H2O2, as second messenger. Then change the redox state in the plants. Monomer NPR1 (nonexpressor of pathogenesis-related genes 1) activites PR (pathogenesis-related proteins) gene, so plants acquire SAR (systemic acquired resistance) (Lee et al., 2010; Rajjou et al., 2006). Exogenous SA treatment can also activate PR gene, inducing plants acquiring SAR. It shows that exogenous SA can not only make plants respond to biological stress, but also is a signal molecule that induces SAR. This conclusion has been confirmed in tobacco, cucumber and Arabidopsis. The role of SA in plant growth and development is still controversial (Gao et al., 2014). Especially in the identification and characteristics of SA receptor, there is no specific receptor reported at present (Mariana and Javier, 2011).
Studies have shown that exogenous SA inhibited seed germination. Such as SA doses> 1mM delays or even suppresses Arabidopsis seed germination (Rajjou et al., 2006). Doses >0.25 mM SA inhibit barley seed germination. While SA concentrations ranging from 3 mM to 5 mM completely inhibit maize germination. This may be due to the addition of exogenous SA induces plant producing oxidative stress and improving copper, zinc superoxide dismutase enzyme activity, as a result of hydrogen peroxide degrading enzymes, catalase and ascorbate peroxidase inactivated, and leading to an increased level of H2O2 in plants, thereby inhibiting seed germination (Mariana and Javier, 2011). The detailed mechanism of SA in seed germination is still unclear.
The conclusion shows that AtR8 RNA is hardly or trace expressed in Arabidopsis dry seeds, but largely existed in the germination seeds. The deletion of AtR8 RNA reduces Arabidopsis seed germination rate under normal culture condition. Under SA stress treatment, the expression of AtR8 RNA is induced, and the deletion of AtR8 RNA further reduces the Arabidopsis seed germination rate. We speculate AtR8 RNA may lie on lncRNA, and affect Arabidopsis seed germination by oxidative stress or SA signal transduction pathway, but the exact mechanism remains unclear.
Acknowledgments
This work was supported by the Program for the Central College Fundamental Research Funds of Northeast Forestry University (DL12BA38), the Central College Fundamental Research Funds Northeast Forestry University (DL13EA04-02), Innovative Research Team of the Ministry of Education (IRT13053) and Scientific Research Foundation for Returned Scholars of the Ministry of Education (47th Batch).
Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Molecular Cell, 2014, 55(3):383-396.
http://dx.doi.org/10.1016/j.molcel.2014.06.011
Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JW, Crespi M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Developmental Cell, 2014, 30(2):166–176.
http://dx.doi.org/10.1016/j.devcel.2014.06.017
Ben Amor B, Wirth S, Merchan F, Laporte P, d'Aubenton-Carafa Y, Hirsch J, Maizel A, Mallory A, Lucas A, Deragon JM, Vaucheret H, Thermes C, Crespi M. Novel long non-protein coding RNAs involved in arabidopsis differentiation and stress responses. Genome research, 2009, 19 (1): 57-69.
http://dx.doi.org/10.1101/gr.080275.108
Chen F, Dahal P, Bradford KJ. Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiology, 2001, 127(3):928-36.
http://dx.doi.org/10.1104/pp.010259
Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci US A, 2012,109(7): 2654-2659.
http://dx.doi.org/10.1073/pnas.1121374109
Gao Qixin, Hu Xinxi, Wang Huanyan, Cao Ke, Suo Huan, He Changzheng, Research progress in mechanism of plant disease resistance induced by salicylic acid. Chinese Potato Journal, 2014(04): 238 – 242.
Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell, 1992, 4(10): 1251-1261.
http://dx.doi.org/10.2307/3869411
Heo, J.B., S. Sung. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA, Science, 2011, 331(6013): 76-79.
http://dx.doi.org/10.1126/science.1197349
Holdsworth MJ, Bentsink L, Soppe WJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol, 2008, 179(1): 33-54.
http://dx.doi.org/10.1111/j.1469-8137.2008.02437.x
Huang XQ, Li DD, Wu J. Long non-coding RNAs in plants. Hereditas (Beijing), 2015, 37(4): 344-359.
Kovácik J, Grúz J, Backor M, Strnad M, Repcák M. Salicylic acid-induced changes to growth and phenolic metabolism in Matricaria chamomilla plants. Plant cell reports, 2009, 28(1): 135-143.
http://dx.doi.org/10.1007/s00299-008-0627-5
Krishnan P., J.D.K., Shantha Nagarajan, Moharir A.V. Characterization of germinating and non-viable soybean seeds by nuclear magnetic resonance (NMR) spectroscopy. Seed Science Research, 2004, 14(4): 355-362.
http://dx.doi.org/10.1079/SSR2004189
Lee S, Kim SG, Park CM. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytologist, 2010, 188(2): 626-637.
http://dx.doi.org/10.1111/j.1469-8137.2010.03378.x
Mariana Rivas-San Vicente and Javier Plasencia. Salicylic acid beyond defence: its role in plant growth and development. Journal of Experimental Botany, 2011, 62(10):3321-3338.
http://dx.doi.org/10.1093/jxb/err031
Martínez C, Pons E, Prats G, León J. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant Journal for Cell & Molecular Biology, 2004, 37(2):209–217.
http://dx.doi.org/10.1046/j.1365-313X.2003.01954.x
Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T. Analysis of ABA Hypersensitive Germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant Journal for Cell & Molecular Biology, 2005, 44(6):972-984.
http://dx.doi.org/10.1111/j.1365-313X.2005.02589.x
Po-Pu Liu, TaiowaA.Montgomery, Noah Fahlgren, Kristin D. Kasschau, Hiroyuki Nonogaki, James C. Carrington. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. The Plant Journal, 2007, 52(1): 133-146.
http://dx.doi.org/10.1111/j.1365-313X.2007.03218.x
Rajjou L, Belghazi M, Huguet R, Robin C, Moreau A, Job C, Job D. Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiology, 2006,141(3): 910–923.
http://dx.doi.org/10.1104/pp.106.082057
Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets, Cell, 2002, 110(4): 513-520.
http://dx.doi.org/10.1016/S0092-8674(02)00863-2
Ruth C. Martin, Masashi Asahina, Po-Pu Liu, Jessica R. Kristof,Jennifer L. Coppersmith, Wioletta E. Pluskota, George W. Bassel,Natalya A. Goloviznina, Theresa T. Nguyen, Cristina Martı´nez-Andu´ jar,M.B. Arun Kumar, Piotr Pupel, Hiroyuki Nonogaki. The microRNA156 and microRNA 172 gene regulation cascades at post-germinative stages in Arabidopsis, Seed Science Research, 2010, 20(2):79-87.
Ruth C. Martin, Masashi Asahina, Po-Pu Liu, Jessica R. Kristof,Jennifer L. Coppersmith, Wioletta E. Pluskota, George W. Bassel, Natalya A. Goloviznina, Theresa T. Nguyen, Cristina Martı´nez-Andu´ jar,M.B. Arun Kumar, Piotr Pupel, Hiroyuki Nonogaki. The regulation of post-germinative transition from the cotyledonto vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis, Seed Science Research, 2010, 20(2): 89-96.
http://dx.doi.org/10.1017/S0960258510000073
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal, 1995, 8(5): 659-671.
http://dx.doi.org/10.1046/j.1365-313X.1995.08050659.x
Swiezewski S, Liu F, Magusin A, Dean C, Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target, Nature, 2009, 462(7274): 799-802.
http://dx.doi.org/10.1038/nature08618
Wang Fang, Pang Zixue, Wang Hanning, Mu Yanzhao, Li Hongbin. Effects of Salicylic Acid Pretreatment on the Seeds Germination and Physiological Characteristics of Sweet Corn, Journal of Maize Sciences, 2012, 20(4): 74-77.
Wang Y, Fan X, Lin F, He G, Terzaghi W, Zhu D, Deng XW. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(28):10359-64.
http://dx.doi.org/10.1073/pnas.1409457111
Wu J, Toshihiro Okada, Toru Fukushima, Takahiko Tsudzuki, Masahiro Sugiura, Yasushi Yukawa. A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biology, 2012, 9(3): 302-313.
http://dx.doi.org/10.4161/rna.19101
Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS One, 2014, 9(1): e85208.
http://dx.doi.org/10.1371/journal.pone.0085208
Zhang J, M.H., Hou YX, Nallamilli BR, Peng ZH. Plant Long ncRNAs: A new frontier for gene regulatory control. American Journal of Plant Sciences, 2013, 04(05):1038-1045.
. PDF(218KB)
. FPDF(win)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Dandan Li
. Xiaoqing Huang
. Ziguang Liu
. Shuang Li
. Toshihiro Okada
. Yasushi Yukawa
. Juan Wu
Related articles
. Long noncoding RNA
. Arabidopsis
. Seed germination
. Environmental stress response
Tools
. Email to a friend
. Post a comment


